Author: Lukas Hörtnagl (holukas@ethz.ch)
Imports#
import importlib.metadata
import warnings
from datetime import datetime
from pathlib import Path
import pandas as pd
import matplotlib.pyplot as plt
import diive as dv
from diive.core.io.files import save_parquet, load_parquet
from diive.core.plotting.cumulative import CumulativeYear
warnings.filterwarnings(action='ignore', category=FutureWarning)
warnings.filterwarnings(action='ignore', category=UserWarning)
version_diive = importlib.metadata.version("diive")
print(f"diive version: v{version_diive}")
diive version: v0.87.0
Load data#
SOURCEDIR = r"../80_FINALIZE"
FILENAME = r"81.1_FLUXES_M15_MGMT_L4.2_NEE_GPP_RECO_LE_H_FN2O_FCH4.parquet"
FILEPATH = Path(SOURCEDIR) / FILENAME
df = load_parquet(filepath=FILEPATH)
df
Loaded .parquet file ..\80_FINALIZE\81.1_FLUXES_M15_MGMT_L4.2_NEE_GPP_RECO_LE_H_FN2O_FCH4.parquet (1.005 seconds).
--> Detected time resolution of <30 * Minutes> / 30min
| .PREC_RAIN_TOT_GF1_0.5_1_MEAN3H-12 | .PREC_RAIN_TOT_GF1_0.5_1_MEAN3H-18 | .PREC_RAIN_TOT_GF1_0.5_1_MEAN3H-24 | .PREC_RAIN_TOT_GF1_0.5_1_MEAN3H-6 | .SWC_GF1_0.15_1_gfXG_MEAN3H-12 | .SWC_GF1_0.15_1_gfXG_MEAN3H-18 | .SWC_GF1_0.15_1_gfXG_MEAN3H-24 | .SWC_GF1_0.15_1_gfXG_MEAN3H-6 | .TS_GF1_0.04_1_gfXG_MEAN3H-12 | .TS_GF1_0.04_1_gfXG_MEAN3H-18 | .TS_GF1_0.04_1_gfXG_MEAN3H-24 | .TS_GF1_0.04_1_gfXG_MEAN3H-6 | .TS_GF1_0.15_1_gfXG_MEAN3H-12 | .TS_GF1_0.15_1_gfXG_MEAN3H-18 | .TS_GF1_0.15_1_gfXG_MEAN3H-24 | ... | GPP_NT_CUT_50_gfRF | RECO_DT_CUT_50_gfRF | GPP_DT_CUT_50_gfRF | RECO_DT_CUT_50_gfRF_SD | GPP_DT_CUT_50_gfRF_SD | G_GF1_0.03_1 | G_GF1_0.03_2 | G_GF1_0.05_1 | G_GF1_0.05_2 | G_GF4_0.02_1 | G_GF5_0.02_1 | LW_OUT_T1_2_1 | NETRAD_T1_2_1 | PPFD_OUT_T1_2_2 | SW_OUT_T1_2_1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIMESTAMP_MIDDLE | |||||||||||||||||||||||||||||||
| 2005-01-01 00:15:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.918553 | 0.093071 | 0.0 | 0.080016 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2005-01-01 00:45:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.917972 | 0.092682 | 0.0 | 0.079688 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2005-01-01 01:15:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.163001 | 0.093071 | 0.0 | 0.080016 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2005-01-01 01:45:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.190890 | 0.093071 | 0.0 | 0.080016 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2005-01-01 02:15:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.167042 | 0.092295 | 0.0 | 0.079361 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2024-12-31 21:45:00 | 0.0 | 0.0 | 0.0 | 0.0 | 52.229004 | 52.226300 | 52.226689 | 52.216796 | 3.458828 | 3.150402 | 3.115260 | 3.660897 | 4.335667 | 4.347764 | 4.385967 | ... | -0.334996 | 1.091028 | 0.0 | 0.265808 | 0.0 | NaN | NaN | -9.097370 | -7.880106 | NaN | NaN | 311.167160 | -5.883538 | 0.0 | 0.0 |
| 2024-12-31 22:15:00 | 0.0 | 0.0 | 0.0 | 0.0 | 52.227858 | 52.227986 | 52.224528 | 52.214211 | 3.522570 | 3.187638 | 3.103440 | 3.643396 | 4.338551 | 4.342880 | 4.379524 | ... | -0.310533 | 1.078751 | 0.0 | 0.264327 | 0.0 | NaN | NaN | -9.561669 | -8.172388 | NaN | NaN | 310.079817 | -6.269816 | 0.0 | 0.0 |
| 2024-12-31 22:45:00 | 0.0 | 0.0 | 0.0 | 0.0 | 52.226640 | 52.229837 | 52.222456 | 52.209876 | 3.578745 | 3.230037 | 3.095339 | 3.624025 | 4.343767 | 4.339440 | 4.372636 | ... | -0.225651 | 1.079759 | 0.0 | 0.264447 | 0.0 | NaN | NaN | -10.138718 | -8.527732 | NaN | NaN | 309.604987 | -6.934394 | 0.0 | 0.0 |
| 2024-12-31 23:15:00 | 0.0 | 0.0 | 0.0 | 0.0 | 52.224375 | 52.231151 | 52.221324 | 52.238293 | 3.624160 | 3.278488 | 3.093806 | 3.601135 | 4.350872 | 4.336333 | 4.366082 | ... | -0.558285 | 1.062164 | 0.0 | 0.262373 | 0.0 | NaN | NaN | -10.649611 | -8.871628 | NaN | NaN | 308.812117 | -5.696729 | 0.0 | 0.0 |
| 2024-12-31 23:45:00 | 0.0 | 0.0 | 0.0 | 0.0 | 52.222007 | 52.230632 | 52.222701 | 52.273511 | 3.656167 | 3.331678 | 3.103003 | 3.579020 | 4.360311 | 4.334225 | 4.359530 | ... | -0.317543 | 1.047483 | 0.0 | 0.260688 | 0.0 | NaN | NaN | -10.944774 | -9.138224 | NaN | NaN | 307.372117 | -8.102484 | 0.0 | 0.0 |
350640 rows × 812 columns
List of gap-filled flux variables#
fluxlist = [c for c in df.columns if str(c).endswith("gfRF")];
fluxlist
['NEE_L3.1_L3.3_CUT_16_QCF_gfRF',
'NEE_L3.1_L3.3_CUT_50_QCF_gfRF',
'NEE_L3.1_L3.3_CUT_84_QCF_gfRF',
'LE_L3.1_L3.3_CUT_NONE_QCF_gfRF',
'H_L3.1_L3.3_CUT_NONE_QCF_gfRF',
'FN2O_L3.1_L3.3_CUT_16_QCF_gfRF',
'FN2O_L3.1_L3.3_CUT_50_QCF_gfRF',
'FN2O_L3.1_L3.3_CUT_84_QCF_gfRF',
'FCH4_L3.1_L3.3_CUT_16_QCF_gfRF',
'FCH4_L3.1_L3.3_CUT_50_QCF_gfRF',
'FCH4_L3.1_L3.3_CUT_84_QCF_gfRF',
'RECO_NT_CUT_16_gfRF',
'GPP_NT_CUT_16_gfRF',
'RECO_DT_CUT_16_gfRF',
'GPP_DT_CUT_16_gfRF',
'RECO_NT_CUT_84_gfRF',
'GPP_NT_CUT_84_gfRF',
'RECO_DT_CUT_84_gfRF',
'GPP_DT_CUT_84_gfRF',
'RECO_NT_CUT_50_gfRF',
'GPP_NT_CUT_50_gfRF',
'RECO_DT_CUT_50_gfRF',
'GPP_DT_CUT_50_gfRF']
Fluxes: gap-filled (random forest)#
Main ecosystem fluxes#
nee = 'NEE_L3.1_L3.3_CUT_50_QCF_gfRF'
le = 'LE_L3.1_L3.3_CUT_NONE_QCF_gfRF'
h = 'H_L3.1_L3.3_CUT_NONE_QCF_gfRF'
n2o = 'FN2O_L3.1_L3.3_CUT_50_QCF_gfRF'
ch4 = 'FCH4_L3.1_L3.3_CUT_50_QCF_gfRF'
gpp = 'GPP_DT_CUT_50_gfRF'
reco = 'RECO_DT_CUT_50_gfRF'
fluxes = [nee, gpp, reco, le, h, n2o, ch4]
fluxes
['NEE_L3.1_L3.3_CUT_50_QCF_gfRF',
'GPP_DT_CUT_50_gfRF',
'RECO_DT_CUT_50_gfRF',
'LE_L3.1_L3.3_CUT_NONE_QCF_gfRF',
'H_L3.1_L3.3_CUT_NONE_QCF_gfRF',
'FN2O_L3.1_L3.3_CUT_50_QCF_gfRF',
'FCH4_L3.1_L3.3_CUT_50_QCF_gfRF']
Flux units: $\( \mathrm{NEE=\mu mol\ CO_{2}\ m^{-2}\ s^{-1}} \)\( \)\( \mathrm{LE=W\ m^{-2}} \)\( \)\( \mathrm{H=W\ m^{-2}} \)\( \)\( \mathrm{N_{2}O=nmol\ N_{2}O\ m^{-2}\ s^{-1}} \)\( \)\( \mathrm{CH_{4}=nmol\ CH_{4}\ m^{-2}\ s^{-1}} \)$
units_nee = r"$\mathrm{\mu mol\ CO_{2}\ m^{-2}\ s^{-1}}$"
units_gpp = r"$\mathrm{\mu mol\ CO_{2}\ m^{-2}\ s^{-1}}$"
units_reco = r"$\mathrm{\mu mol\ CO_{2}\ m^{-2}\ s^{-1}}$"
units_le = r"$\mathrm{W\ m^{-2}}$"
units_h = r"$\mathrm{W\ m^{-2}}$"
units_n2o = r"$\mathrm{nmol\ N_{2}O\ m^{-2}\ s^{-1}}$"
units_ch4 = r"$\mathrm{nmol\ CH_{4}\ m^{-2}\ s^{-1}}$"
Heatmaps (half-hourly fluxes)#
fig, axs = plt.subplots(ncols=7, figsize=(30, 15), dpi=70, layout="constrained")
dv.heatmapdatetime(series=df[nee], cb_digits_after_comma=0, ax=axs[0], zlabel=units_nee).plot()
dv.heatmapdatetime(series=df[gpp], cb_digits_after_comma=0, ax=axs[1], zlabel=units_gpp).plot()
dv.heatmapdatetime(series=df[reco], cb_digits_after_comma=0, ax=axs[2], zlabel=units_reco).plot()
dv.heatmapdatetime(series=df[le], cb_digits_after_comma=0, ax=axs[3], zlabel=units_le).plot()
dv.heatmapdatetime(series=df[h], cb_digits_after_comma=0, ax=axs[4], zlabel=units_h).plot()
dv.heatmapdatetime(series=df[n2o], cb_digits_after_comma=0, ax=axs[5], zlabel=units_n2o).plot()
dv.heatmapdatetime(series=df[ch4], cb_digits_after_comma=0, ax=axs[6], zlabel=units_ch4).plot()
hide_labels = [1, 2, 3, 4, 5, 6]
for h in hide_labels:
axs[h].axes.get_yaxis().get_label().set_visible(False)
plt.setp(axs[h].get_yticklabels(), visible=False)
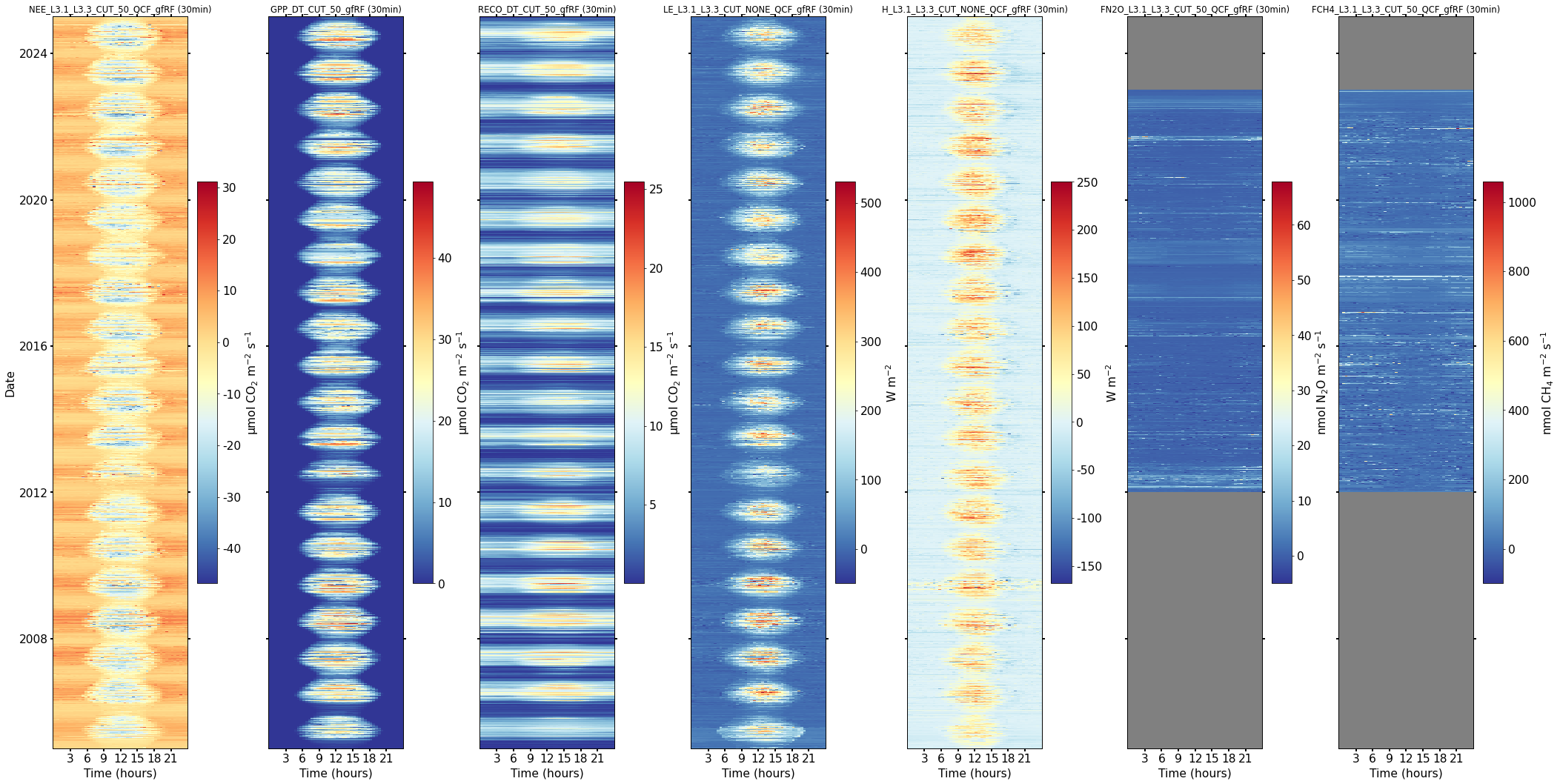
Heatmaps (monthly fluxes)#
fig, axs = plt.subplots(ncols=7, figsize=(36, 12), dpi=150, layout="constrained")
fig.suptitle(f'Mean flux per month', fontsize=32)
for ix, f in enumerate(fluxes):
s = df[f].resample('M').mean()
dv.heatmapyearmonth(series_monthly=s, title=s.name, ax=axs[ix], cb_digits_after_comma=0, zlabel="mean flux").plot()
# axs[0].axes.get_yaxis().get_label().set_visible(False)
hide_labels = [1, 2, 3, 4]
for h in hide_labels:
axs[h].axes.get_yaxis().get_label().set_visible(False)
plt.setp(axs[h].get_yticklabels(), visible=False)
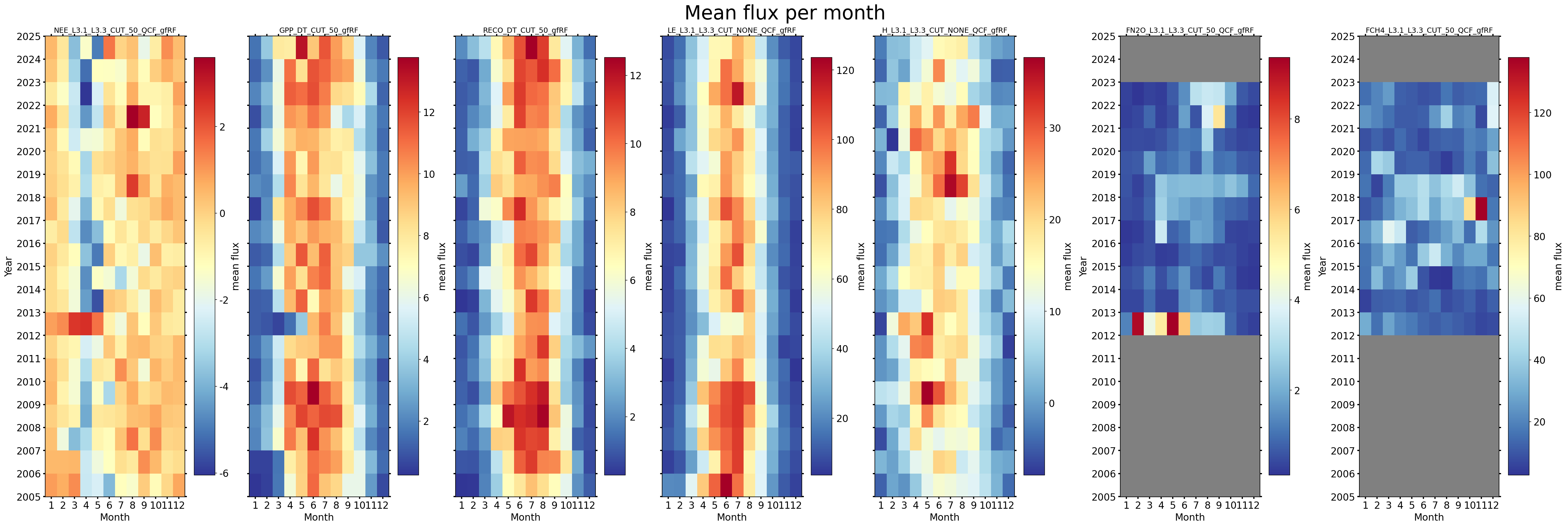
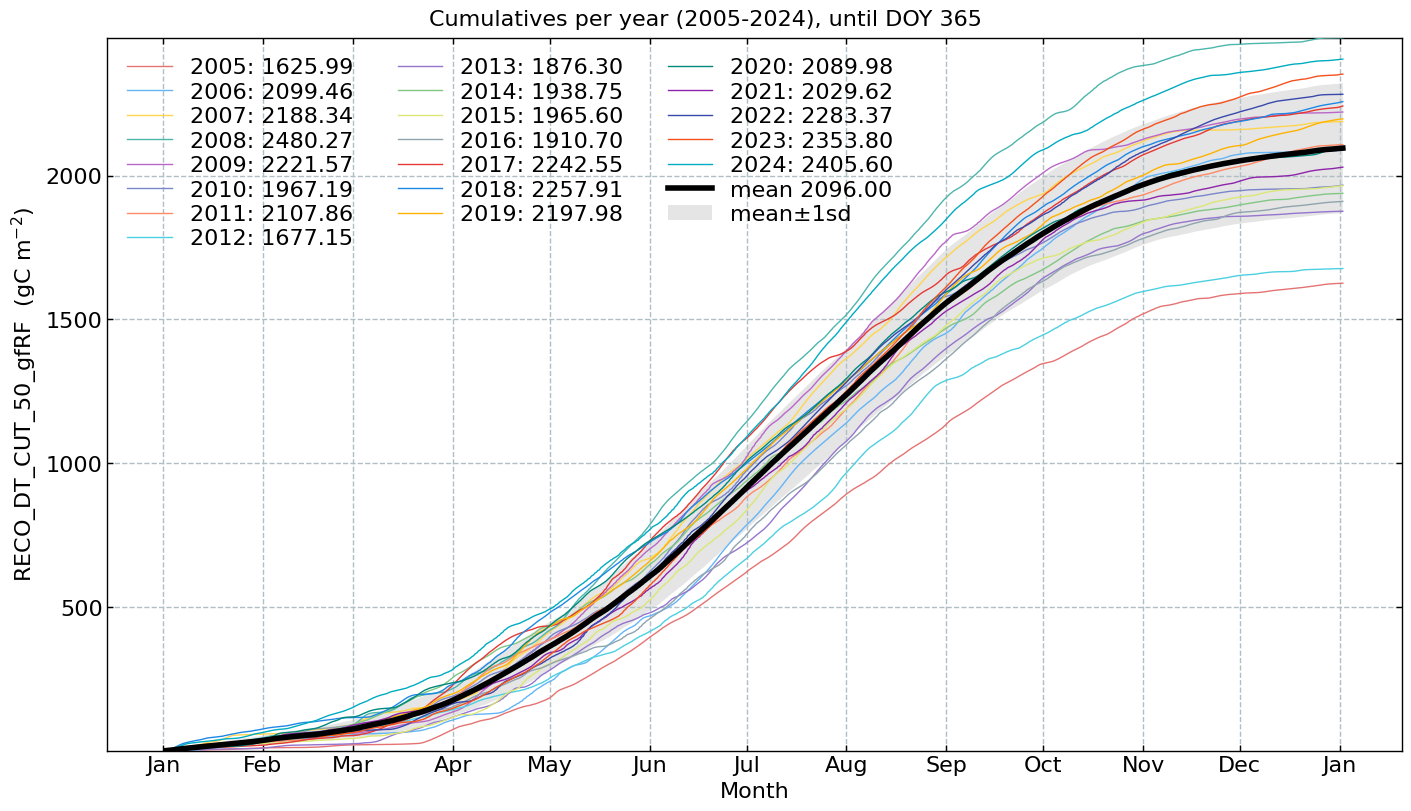
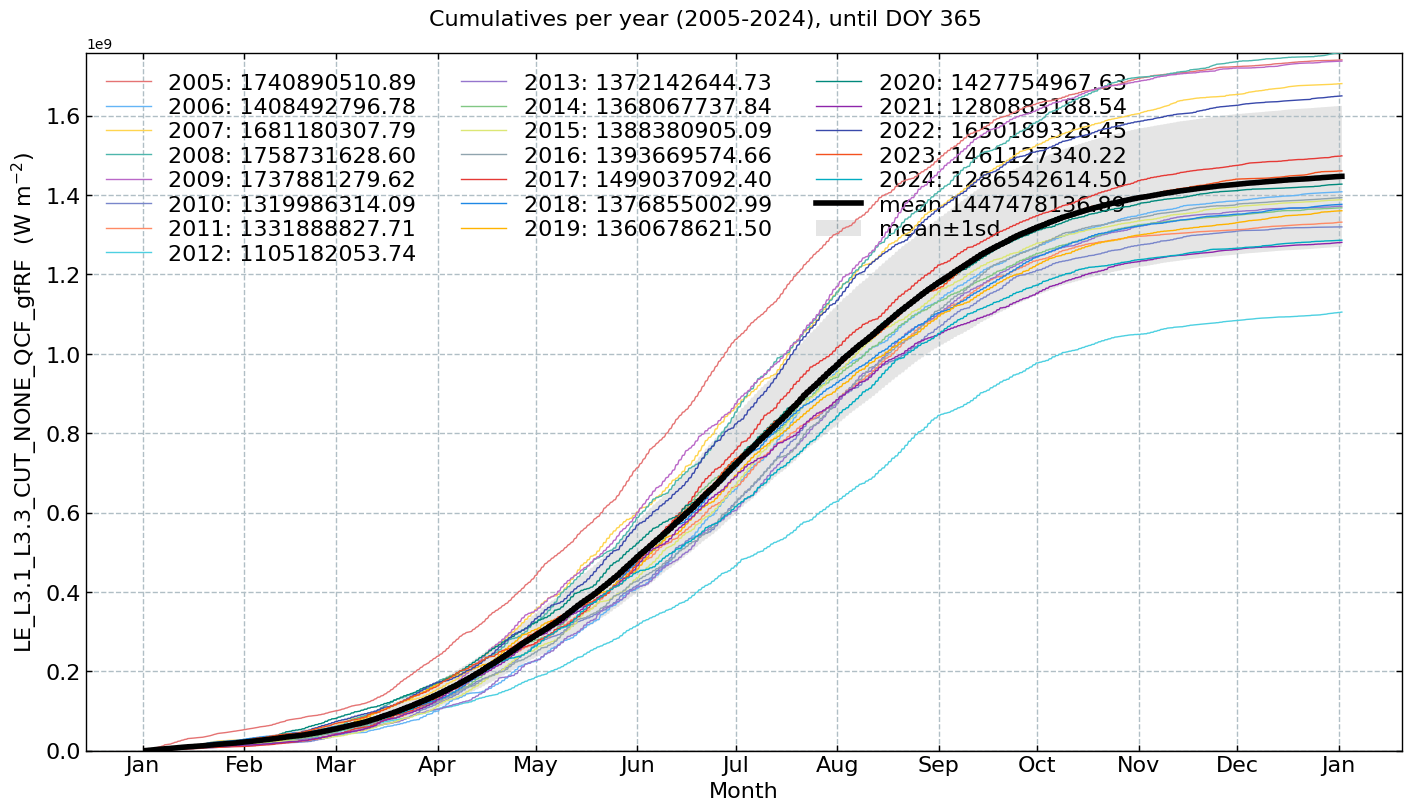
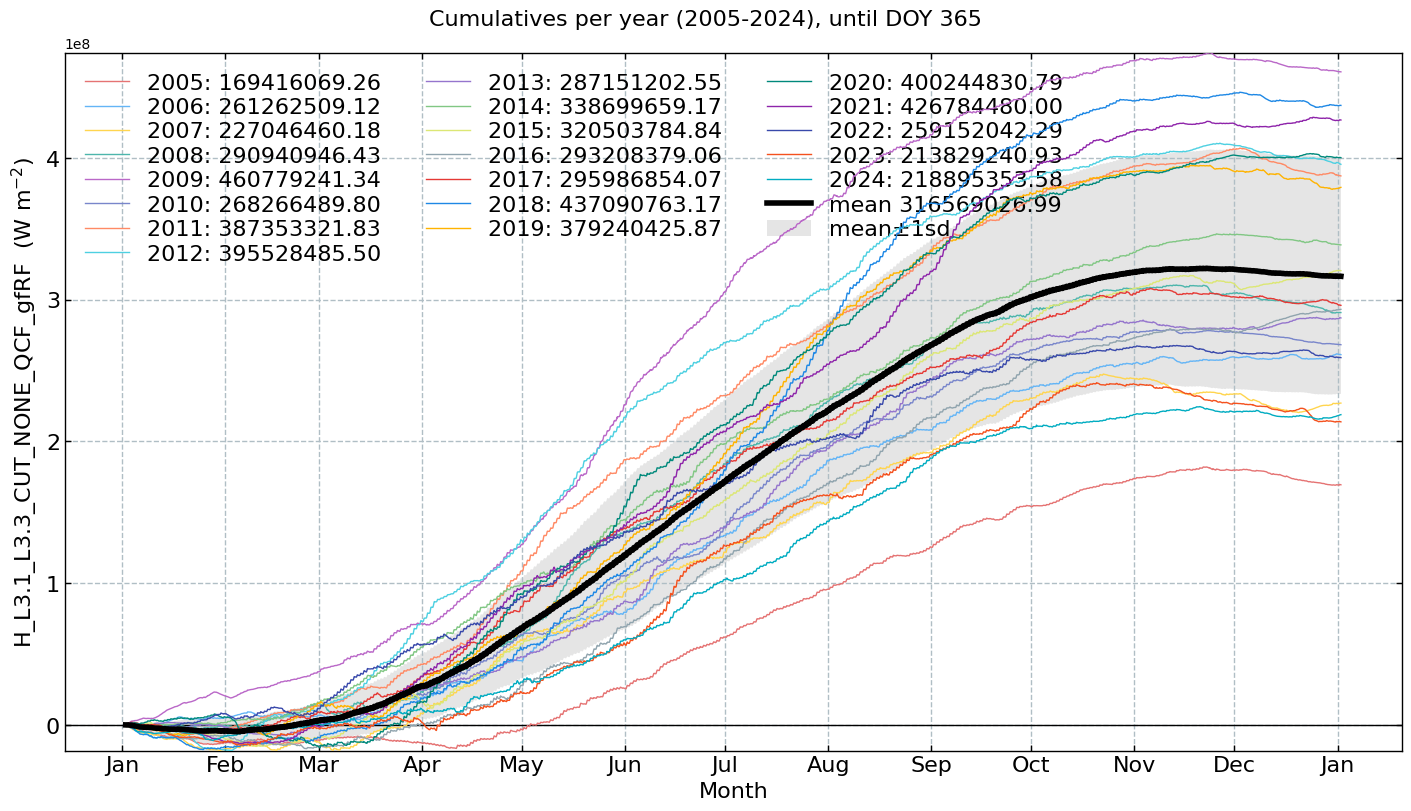
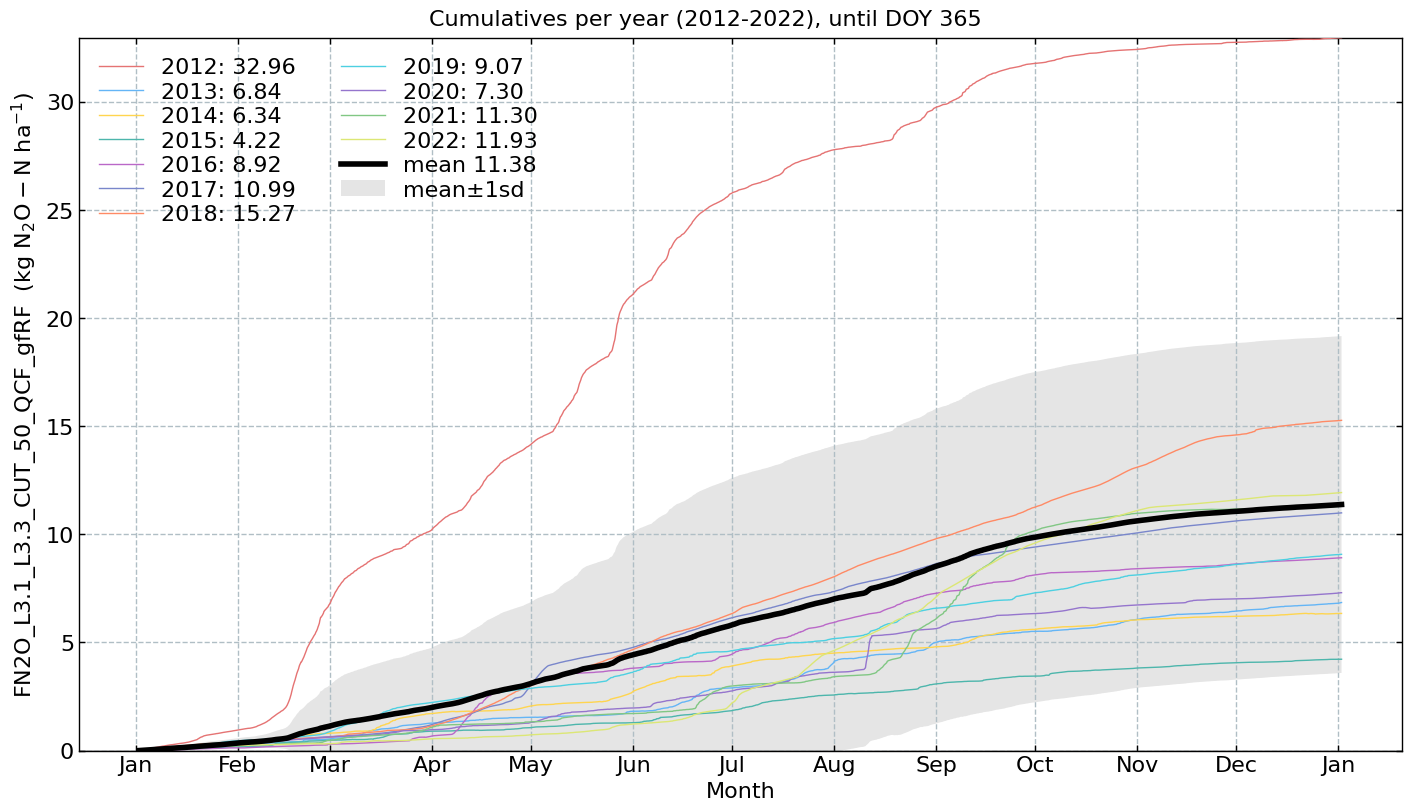
Cumulatives per year#
# Conversion factors and resulting units
conversions = {
'NEE_L3.1_L3.3_CUT_50_QCF_gfRF': [0.02161926, r'($\mathrm{gC\ m^{-2}}$)'], # umol CO2 m-2 s-1 --> gC m-2 30min-1
'GPP_DT_CUT_50_gfRF': [0.02161926, r'($\mathrm{gC\ m^{-2}}$)'], # umol CO2 m-2 s-1 --> gC m-2 30min-1
'RECO_DT_CUT_50_gfRF': [0.02161926, r'($\mathrm{gC\ m^{-2}}$)'], # umol CO2 m-2 s-1 --> gC m-2 30min-1
'LE_L3.1_L3.3_CUT_NONE_QCF_gfRF': [1800, r'($\mathrm{W\ m^{-2}}$)'], # W m-2 s-1 --> W m-2 30min-1
'H_L3.1_L3.3_CUT_NONE_QCF_gfRF': [1800, r'($\mathrm{W\ m^{-2}}$)'], # W m-2 s-1 --> W m-2 30min-1
'FN2O_L3.1_L3.3_CUT_50_QCF_gfRF': [0.00050424109632, r'($\mathrm{kg\ N_2O-N\ ha^{-1}}$)'], # nmol N2O m-2 s-1 --> kg N2O-N ha-1 30min-1
'FCH4_L3.1_L3.3_CUT_50_QCF_gfRF': [0.000216198, r'($\mathrm{kg\ CH_4-C\ ha^{-1}}$)'] # nmol CH4 m-2 s-1 --> kg CH4-C ha-1 30min-1
}
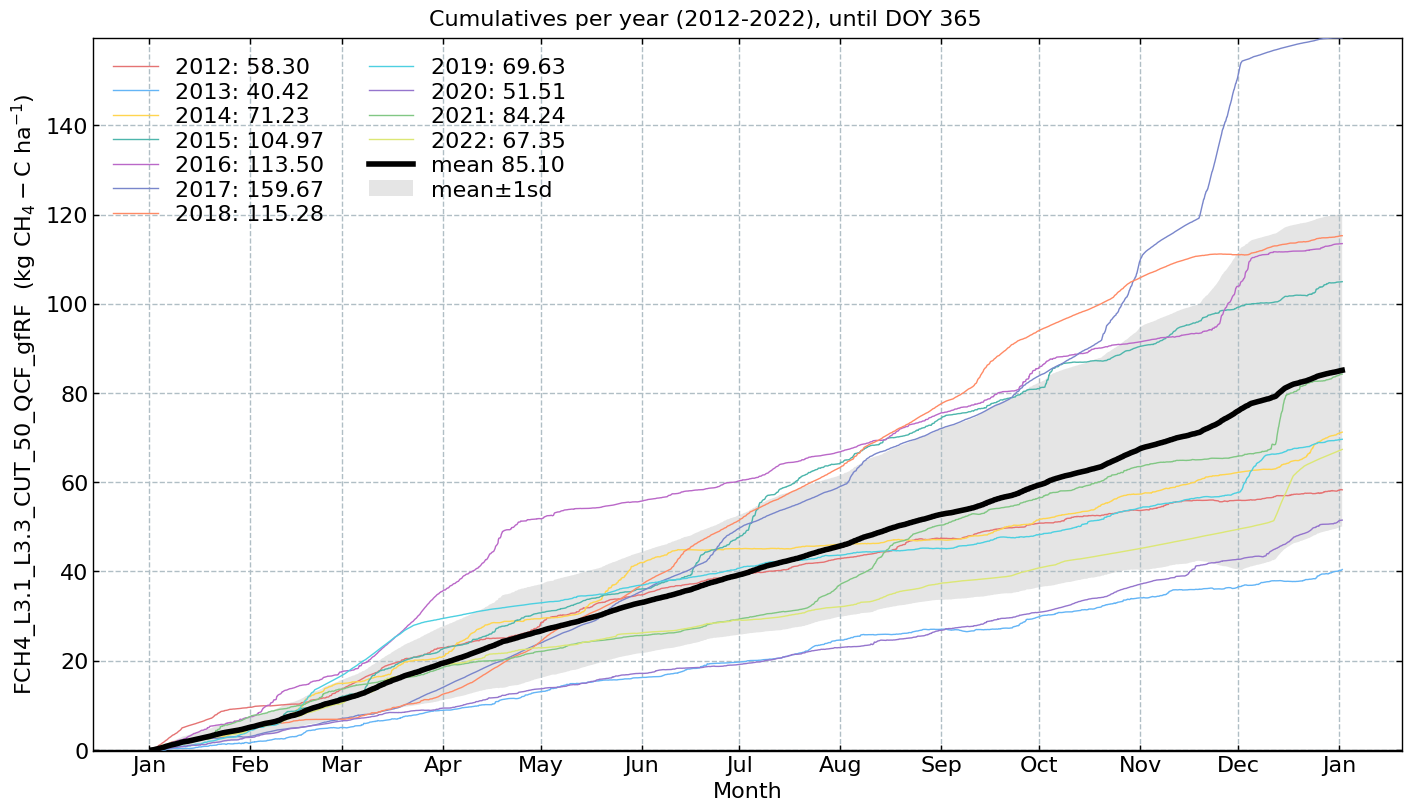
for f in fluxes:
s = df[f].multiply(conversions[f][0])
units = conversions[f][1]
CumulativeYear(
series=s,
series_units=units,
start_year=2005,
end_year=2024,
show_reference=True,
excl_years_from_reference=None,
highlight_year=None,
highlight_year_color='#F44336').plot();
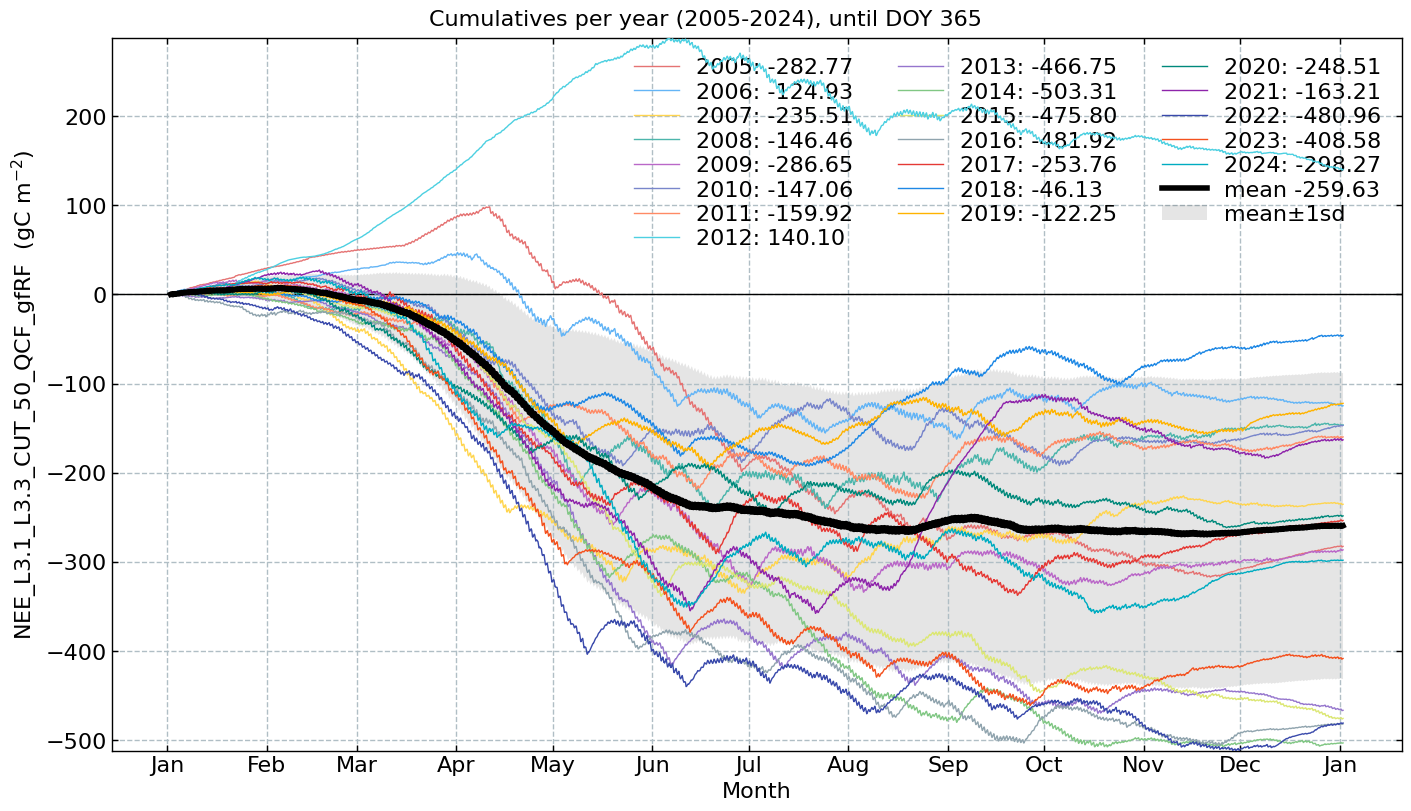
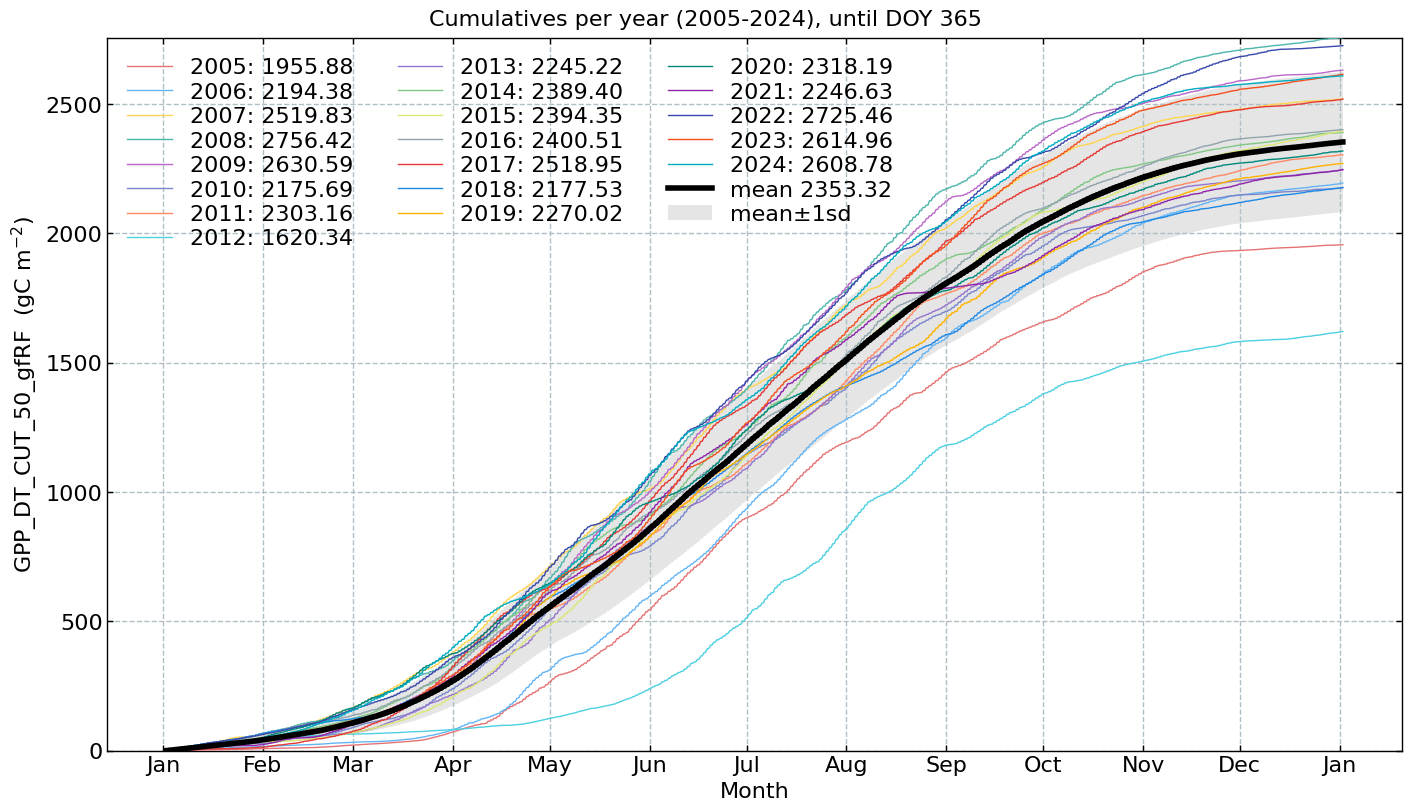
End of notebook#
dt_string = datetime.now().strftime("%Y-%m-%d %H:%M:%S")
print(f"Finished. {dt_string}")
Finished. 2025-05-16 16:02:48
